After a month-long hiatus, thought I'd return with an exposition of a very, very neat paper I read recently. This takes us into the still-inchoate world of computational biology, at the intersection of the platonic mathemtics of computer algorithms and the messy junkyard of cell biology. If you're not versed in the language, try to hang in there, because it's all rather interesting.
The paper is
Network motifs in the transcriptional regulation network of E. coli, by Shai Shen-Orr, Ron Milo, Shmoolik Mangan, and Uri Alon at the Weizmann Institute in Israel. It was published less than three years ago in
Nature, and has already been cited by more than 150 other papers; clearly, the ideas it introduced have caught the imaginations of other researchers.
Before jumping into the paper though, let's begin by framing what a network is.
Networks
A network (also called a graph) is a collection of two things:
1.
nodes, which are some entity that you care about, like people, places, times, or genes.
2.
edges, which express some relationship between nodes. Edges can express a symmetric relation such as "are friends" or "are neighbors", or a one-directional relationship such as "loves", "hates", or "is built from." We call a network with symmetric relations "undirected networks", and the other a "directed network" (or graph).
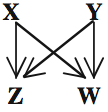
For example, using characters as our nodes, and love-relationships as our edges, we can draw a small directed network representing the love triangle in Shakespeare's
Twelfth Night:
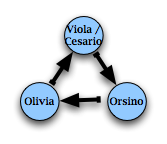
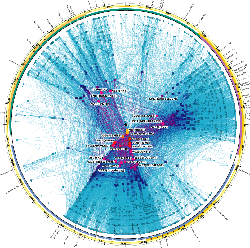
Or, more prosaically, researchers have built a network representing the
core connectivity of the Internet:
 Random Networks
Random Networks
When scientists wanted to study the behavior of networks, they traditionally assumed that a typical
"random graph" had characteristics like a Erdös-Rényi graph, a theoretical construct invented by a pair of fine Hungarian mathematicians; essentially, you start with a bunch of nodes, then flip a (weighted) coin to decide whether to draw an edge connecting each pair of nodes. This process produces a network that looks something like this (
figures from Barabási & Oltvai 2004):
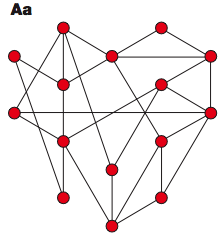
If you make a plot of the
degree distribution, i.e. how many nodes have 1 edge connected, how many have 2 edges, etc., you get a histogram that looks like this for an Erdös-Rényi random graph:
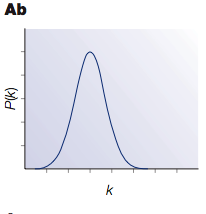
So the principle characteristic of these Erdös-Rényi random graphs is that most nodes have roughly the same number of edges, equal to the average number of edges per node. These networks are fairly uniform-looking; most nodes are about the same as most other nodes. As a real-life example of such a network, you might imagine people sending a chain letter in a small town; most people with send it to five others, but some might send it to more or fewer.
Scale-free networks
Now this is all very well, but when people looked more closely, they began to realize that many real-life networks do
not have the characteristics of these Erdös-Rényi random graphs. Rather than having a whole bunch of nodes that are roughly equally likely to have connecting edges, many of these networks are highly non-uniform: they have a small number of highly connected nodes, and a very large number of nodes that just have one or two edges. A typical scale-free network might look like this:
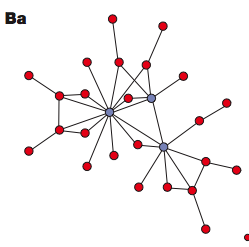
Can you see the "important nodes" in blue? Sometimes those are called "hubs", just like with "hub cities" for the networks of airlines. And this is the corresponding degree distribution:
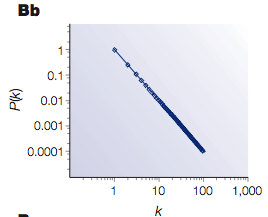
This peculiar degree distribution gives these networks the name
scale-free, for there is no "typical" node, and the degree distributions follow a power law; there are exponentially more nodes with a small number of edges than those with a great many edges.
The Web
has been found to be a scale-free network; there are a few pages that have a great many links, and a vast number of pages with very few links. (The connectivity of Internet routers shares this property as well.) And perhaps thanks to the Hollywood star system, the
network of actors co-starring in movies in the IMDB is also scale-free.
It turns out that many biological networks are also scale-free; the neurons of
C. elegans, the gene regulatory network of
E. coli, protein-protein interaction networks in yeast, among many others, are all scale-free.
Scale-free networks can be modeled, or randomly generated by several methods, all different than the procedure described above for producing an Erdös-Rényi random graph. One simple method is to add a new node, and preferentially connect that new node to the more highly connected nodes that already exist. For example, when you create a new web page, you might tend to create links to sites that already have many links to them, because you will have heard about these sites.
Motifs
So terrific: Scientists now have a better model for real-life networks. But is it sufficient to know that these real-life networks are scale-free? Can we do mathematical exploration of the characteristics of random scale-free graphs, and apply the lessons learned to the real-life networks? Or do the real-life networks have further unique characteristics that a simple model doesn't capture?
This is the question that Shen-Orr et al. set out to ask in this interesting paper; the answer, as we shall see, is
yes, real-life networks are not just random scale-free networks.
Shen-Orr chose as their first example the gene regulation network of
E. coli; it's a fairly small and well-understood network, so it was a good first target. In this network, each node represents a gene; an arrow is drawn from gene A to gene B if the protein that A encodes acts to alter the rate of expression of gene B (regardless of whether A upregulates or downregulates B).To answer the question of whether random scale-free networks are a good model of this network, they generated thousands of random networks, each of which had the same degree distribution as the original, real network. That is, each of these networks had 178 nodes with one edge, 54 nodes with two edges, etc., just as the original did, but which actual nodes were connected to which was randomized.
Shen-Orr et al. then searched each of these networks for
subgraphs, which are just networks within the network, of a certain size. For example, in a directed graph (in which the edges are arrows), there are thirteen possible subgraphs of size three:
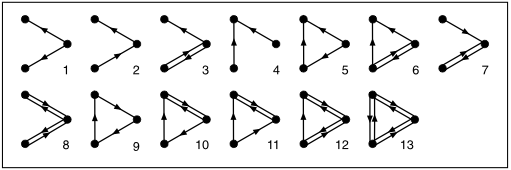
They then compared the frequency of occurrence of each of these types in the
real network and in the many random networks. This is the fascinating result: Of all those 13 subgraphs of size three, one occurred much more frequently in the real network -- number 5 in the diagram.
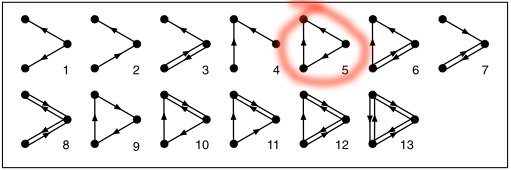
They dubbed this subgraph a "feed-forward loop", because if you arrange its nodes like this:
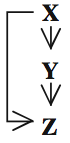
you can imagine gene X exerting influence on gene Z by two pathways: one direct, and one indirect, by regulating gene Y which in turn regulates gene Z.
This result means that there is probably some biologically useful property of this type of subgraph. It has been speculated by others that this type of subgraph is a more stable transmitter of information from gene X to gene Z than other possible arrangements, but the role of the feed-forward loop is not yet clear.
Looking at four-node subgraphs, of which there are 199 possible, it was found that the real network had a much higher-than-random frequency of this subgraph, dubbed the "bi-fan":
Again, work is just beginning on understanding the function of this subgraph, and why the cell apparently considers it so useful.
These unusually common subgraphs are called "motifs", by analogy to the repeated patterns in art, architecture, or gene sequences. Once Shen-Orr et al. prompted people to look for them, people have discovered motifs in all kinds of networks, from a wide spectrum of biological networks, to human-made ones like electrical circuits. Why do biological networks have these motifs? Work is just barely beginning to work out plausible explanations.
One interesting result has to do with a closer look at the "feed-forward loop" motif we saw above. In the context of gene regulation, each edge can either represent a positive or negative effect on the expression of the downstream gene. That means that a feed-forward loop can be one of two types: coherent, or incoherent. A coherent feed-forward loop is one in which the direct path from X to Z has the same net effect as the indirect path; i.e. either both have a positive effect, or both have a negative effect:
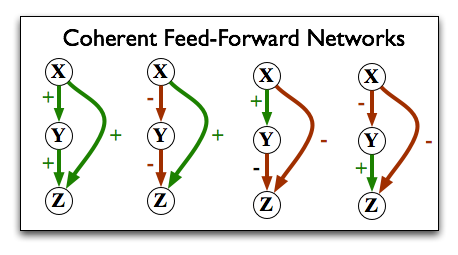
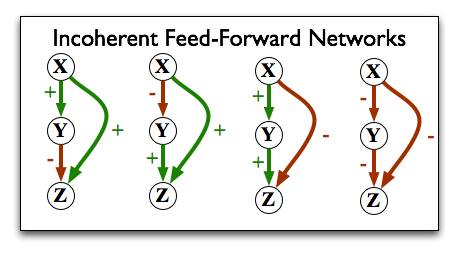
In an incoherent feed-forward loop, the effect from one direction is contradicted by the effect from the other direction:
Shen-Orr et al. found that 85% of the feed-forward loops in the
E. coli regulatory network are coherent. Why should this be? It makes a certain amount of "sense" that you'd want to have the overall regulatory effect of turning on gene X be coherent, but there's still lots to learn. One recent paper out of India has found that theoretically, one particular type of coherent feed-forward loop is a more stable transmitter of information from X to Z; and indeed, that particular type is the most common. So evolution appears to be selecting for good information flow in her networks, perhaps not surprising, but very interesting.
Conclusions
Overall, these results (which have been extended to many other biological and non-biological networks) mean that investigating the mathematical properties of random scale-free networks won't model the behavior of the
E. coli gene regulatory network, or many other biological networks, very well, because they is significantly different than random networks.
Of course, a "random" network is simply one generated by some stochastic process; different processes will produce different types of "random" networks. So far no one has found a simple generative process that produces networks that preferentially contain feedforward-loops and bi-fans, but if someone does, that might yield insights into the function and evolution of biological networks.

 I'm not going to get into the utter tragedy that is this case. I just want to register my confusion and disappointment at this protest imagery.
What is this image supposed to mean?
I'm not going to get into the utter tragedy that is this case. I just want to register my confusion and disappointment at this protest imagery.
What is this image supposed to mean?
 I thought these pro-life types were pretty experienced and savvy protesters, but apparently they need an aesthetic advisor.
Okay, enough snark for the day.
I thought these pro-life types were pretty experienced and savvy protesters, but apparently they need an aesthetic advisor.
Okay, enough snark for the day.
 Or, more prosaically, researchers have built a network representing the
Or, more prosaically, researchers have built a network representing the  Random Networks
When scientists wanted to study the behavior of networks, they traditionally assumed that a typical
Random Networks
When scientists wanted to study the behavior of networks, they traditionally assumed that a typical  If you make a plot of the degree distribution, i.e. how many nodes have 1 edge connected, how many have 2 edges, etc., you get a histogram that looks like this for an Erdös-Rényi random graph:
If you make a plot of the degree distribution, i.e. how many nodes have 1 edge connected, how many have 2 edges, etc., you get a histogram that looks like this for an Erdös-Rényi random graph:
 So the principle characteristic of these Erdös-Rényi random graphs is that most nodes have roughly the same number of edges, equal to the average number of edges per node. These networks are fairly uniform-looking; most nodes are about the same as most other nodes. As a real-life example of such a network, you might imagine people sending a chain letter in a small town; most people with send it to five others, but some might send it to more or fewer.
Scale-free networks
Now this is all very well, but when people looked more closely, they began to realize that many real-life networks do not have the characteristics of these Erdös-Rényi random graphs. Rather than having a whole bunch of nodes that are roughly equally likely to have connecting edges, many of these networks are highly non-uniform: they have a small number of highly connected nodes, and a very large number of nodes that just have one or two edges. A typical scale-free network might look like this:
So the principle characteristic of these Erdös-Rényi random graphs is that most nodes have roughly the same number of edges, equal to the average number of edges per node. These networks are fairly uniform-looking; most nodes are about the same as most other nodes. As a real-life example of such a network, you might imagine people sending a chain letter in a small town; most people with send it to five others, but some might send it to more or fewer.
Scale-free networks
Now this is all very well, but when people looked more closely, they began to realize that many real-life networks do not have the characteristics of these Erdös-Rényi random graphs. Rather than having a whole bunch of nodes that are roughly equally likely to have connecting edges, many of these networks are highly non-uniform: they have a small number of highly connected nodes, and a very large number of nodes that just have one or two edges. A typical scale-free network might look like this:
 Can you see the "important nodes" in blue? Sometimes those are called "hubs", just like with "hub cities" for the networks of airlines. And this is the corresponding degree distribution:
Can you see the "important nodes" in blue? Sometimes those are called "hubs", just like with "hub cities" for the networks of airlines. And this is the corresponding degree distribution:
 This peculiar degree distribution gives these networks the name
This peculiar degree distribution gives these networks the name  They then compared the frequency of occurrence of each of these types in the real network and in the many random networks. This is the fascinating result: Of all those 13 subgraphs of size three, one occurred much more frequently in the real network -- number 5 in the diagram.
They then compared the frequency of occurrence of each of these types in the real network and in the many random networks. This is the fascinating result: Of all those 13 subgraphs of size three, one occurred much more frequently in the real network -- number 5 in the diagram.
 They dubbed this subgraph a "feed-forward loop", because if you arrange its nodes like this:
They dubbed this subgraph a "feed-forward loop", because if you arrange its nodes like this:
 you can imagine gene X exerting influence on gene Z by two pathways: one direct, and one indirect, by regulating gene Y which in turn regulates gene Z.
This result means that there is probably some biologically useful property of this type of subgraph. It has been speculated by others that this type of subgraph is a more stable transmitter of information from gene X to gene Z than other possible arrangements, but the role of the feed-forward loop is not yet clear.
Looking at four-node subgraphs, of which there are 199 possible, it was found that the real network had a much higher-than-random frequency of this subgraph, dubbed the "bi-fan":
you can imagine gene X exerting influence on gene Z by two pathways: one direct, and one indirect, by regulating gene Y which in turn regulates gene Z.
This result means that there is probably some biologically useful property of this type of subgraph. It has been speculated by others that this type of subgraph is a more stable transmitter of information from gene X to gene Z than other possible arrangements, but the role of the feed-forward loop is not yet clear.
Looking at four-node subgraphs, of which there are 199 possible, it was found that the real network had a much higher-than-random frequency of this subgraph, dubbed the "bi-fan":
 Again, work is just beginning on understanding the function of this subgraph, and why the cell apparently considers it so useful.
These unusually common subgraphs are called "motifs", by analogy to the repeated patterns in art, architecture, or gene sequences. Once Shen-Orr et al. prompted people to look for them, people have discovered motifs in all kinds of networks, from a wide spectrum of biological networks, to human-made ones like electrical circuits. Why do biological networks have these motifs? Work is just barely beginning to work out plausible explanations.
One interesting result has to do with a closer look at the "feed-forward loop" motif we saw above. In the context of gene regulation, each edge can either represent a positive or negative effect on the expression of the downstream gene. That means that a feed-forward loop can be one of two types: coherent, or incoherent. A coherent feed-forward loop is one in which the direct path from X to Z has the same net effect as the indirect path; i.e. either both have a positive effect, or both have a negative effect:
Again, work is just beginning on understanding the function of this subgraph, and why the cell apparently considers it so useful.
These unusually common subgraphs are called "motifs", by analogy to the repeated patterns in art, architecture, or gene sequences. Once Shen-Orr et al. prompted people to look for them, people have discovered motifs in all kinds of networks, from a wide spectrum of biological networks, to human-made ones like electrical circuits. Why do biological networks have these motifs? Work is just barely beginning to work out plausible explanations.
One interesting result has to do with a closer look at the "feed-forward loop" motif we saw above. In the context of gene regulation, each edge can either represent a positive or negative effect on the expression of the downstream gene. That means that a feed-forward loop can be one of two types: coherent, or incoherent. A coherent feed-forward loop is one in which the direct path from X to Z has the same net effect as the indirect path; i.e. either both have a positive effect, or both have a negative effect:
 In an incoherent feed-forward loop, the effect from one direction is contradicted by the effect from the other direction:
In an incoherent feed-forward loop, the effect from one direction is contradicted by the effect from the other direction:
 Shen-Orr et al. found that 85% of the feed-forward loops in the E. coli regulatory network are coherent. Why should this be? It makes a certain amount of "sense" that you'd want to have the overall regulatory effect of turning on gene X be coherent, but there's still lots to learn. One recent paper out of India has found that theoretically, one particular type of coherent feed-forward loop is a more stable transmitter of information from X to Z; and indeed, that particular type is the most common. So evolution appears to be selecting for good information flow in her networks, perhaps not surprising, but very interesting.
Conclusions
Overall, these results (which have been extended to many other biological and non-biological networks) mean that investigating the mathematical properties of random scale-free networks won't model the behavior of the E. coli gene regulatory network, or many other biological networks, very well, because they is significantly different than random networks.
Of course, a "random" network is simply one generated by some stochastic process; different processes will produce different types of "random" networks. So far no one has found a simple generative process that produces networks that preferentially contain feedforward-loops and bi-fans, but if someone does, that might yield insights into the function and evolution of biological networks.
Shen-Orr et al. found that 85% of the feed-forward loops in the E. coli regulatory network are coherent. Why should this be? It makes a certain amount of "sense" that you'd want to have the overall regulatory effect of turning on gene X be coherent, but there's still lots to learn. One recent paper out of India has found that theoretically, one particular type of coherent feed-forward loop is a more stable transmitter of information from X to Z; and indeed, that particular type is the most common. So evolution appears to be selecting for good information flow in her networks, perhaps not surprising, but very interesting.
Conclusions
Overall, these results (which have been extended to many other biological and non-biological networks) mean that investigating the mathematical properties of random scale-free networks won't model the behavior of the E. coli gene regulatory network, or many other biological networks, very well, because they is significantly different than random networks.
Of course, a "random" network is simply one generated by some stochastic process; different processes will produce different types of "random" networks. So far no one has found a simple generative process that produces networks that preferentially contain feedforward-loops and bi-fans, but if someone does, that might yield insights into the function and evolution of biological networks.
